Example on computing ionization potential and electron affinity from self-consistent GW for Nitrogen molecule
Here we provide a simple example on how to run Green/Weakcoupling for computing ionization potential (IP) for nitrogen molecule (Geometry obtained for GW100 dataset).
Prerequisites: you must have the mbpt compiled and the green-mbtools python package installed.
Change to a new directory where you will keep your simulations, create a directory for the $N_2$ simulation.
Then create a file with the atom positions in the atom.dat:
N 0.0000 0.0000 0.0000
N 0.0000 0.0000 1.0977The remaining parameters will be specified on the command line.
We then obtain input parameters and the initial mean-field solution by running pySCF via the init_data_mol_df.py script:
python <source root>/green-mbpt/python/init_data_mol_df.py \
--atom atom.dat --basis ccpvdz --xc PBE0We use the ccpvdz basis-set and hybrid PBE0 exchange correlation potential as reference. Not specifying the xc would run Hartree-Fock.
The primary goal of this example is to run a molecular example using fully self-consistent GW and perform analytical continuation to obtain photoelectron spectra and IP.
After that we will run the GW approximation
We then store results into sim.h5 file.
Now we will use the green-mbtools package to perform Nevanlinna analytical continuation from the imaginary axis to real axis.
python <source root>/green-mbtools/examples/nvnl_analysis_mol.py --input input.h5 --sim sim.h5 --iter -1 --beta=1000 \
--grid_file ir/1e5.h5 --out ac_out.h5 \
--e_min -5.0 --e_max 5.0 --n_omega 4000 --eta 0.01This will run Nevanlinna analytical continuation for the data obtained at the last iteration. The output will be stored in the group ac_out.h5.
A plot for the specrta can then be obtained with the plot_dos_mol.py script:
python <source root>/green-mbtools/examples/plot_dos_mol.py --ac_out nvnl_out.h5This will read the analytically continued data and plot it to an <output_dir>/spectra.pdf file. In addition, it will also print the ionization potential (IP) and elecrton affinites (EA).
The resultant spectra looks like:
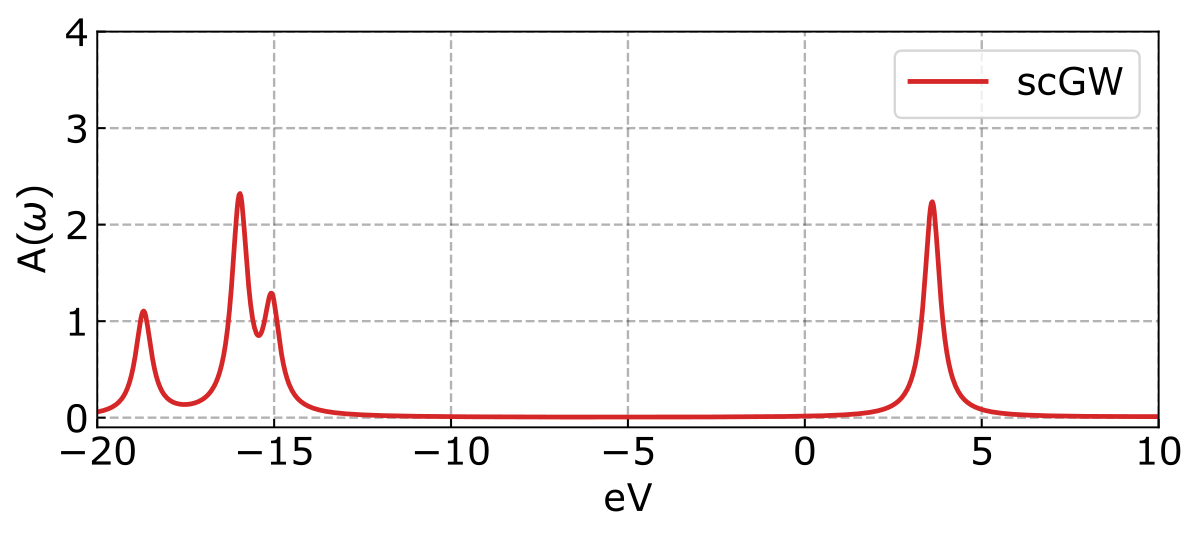
And the ionization potentials (IP) and electron affinities (EA) are displayed as follows.
Lowest 4 Ionization Potentials:
GW IP 1: -15.964 eV
GW IP 2: -15.964 eV
GW IP 3: -15.066 eV
GW IP 4: -18.685 eV
Lowest 4 Electron Affinities:
GW EA 1: 3.604 eV
GW EA 2: 3.604 eV
GW EA 3: 15.143 eV
GW EA 4: 21.321 eVIt is important to highlight that the quasiparticle energies are ordering according to the mean-field reference. As a result, the quasiparticle energies may not be arranged in increasing order because of corrections from dynamic self-energy.